Plot frequencies of multiple categories within a data.frame in
a new fancy way. Tidyverse friendly, based on lares::freqs(),
no limits on amount of features to evaluate.
Usage
freqs_plot(
df,
...,
top = 10,
rm.na = FALSE,
abc = FALSE,
title = NA,
subtitle = NA,
quiet = FALSE
)Arguments
- df
Data.frame
- ...
Variables. Variables you wish to process. Order matters. If no variables are passed, the whole data.frame will be considered
- top
Integer. Filter and plot the most n frequent for categorical values. Set to NA to return all values
- rm.na
Boolean. Remove NA values in the plot? (not filtered for numerical output; use na.omit() or filter() if needed)
- abc
Boolean. Do you wish to sort by alphabetical order?
- title
Character. Overwrite plot's title with.
- subtitle
Character. Overwrite plot's subtitle with.
- quiet
Boolean. Keep quiet? If not, informative messages will be shown.
See also
Other Frequency:
freqs(),
freqs_df(),
freqs_list()
Other Exploratory:
corr_var(),
crosstab(),
df_str(),
distr(),
freqs(),
freqs_df(),
freqs_list(),
lasso_vars(),
missingness(),
plot_cats(),
plot_df(),
plot_nums(),
tree_var()
Other Visualization:
distr(),
freqs(),
freqs_df(),
freqs_list(),
noPlot(),
plot_chord(),
plot_survey(),
plot_timeline(),
tree_var()
Examples
Sys.unsetenv("LARES_FONT") # Temporal
data(dft) # Titanic dataset
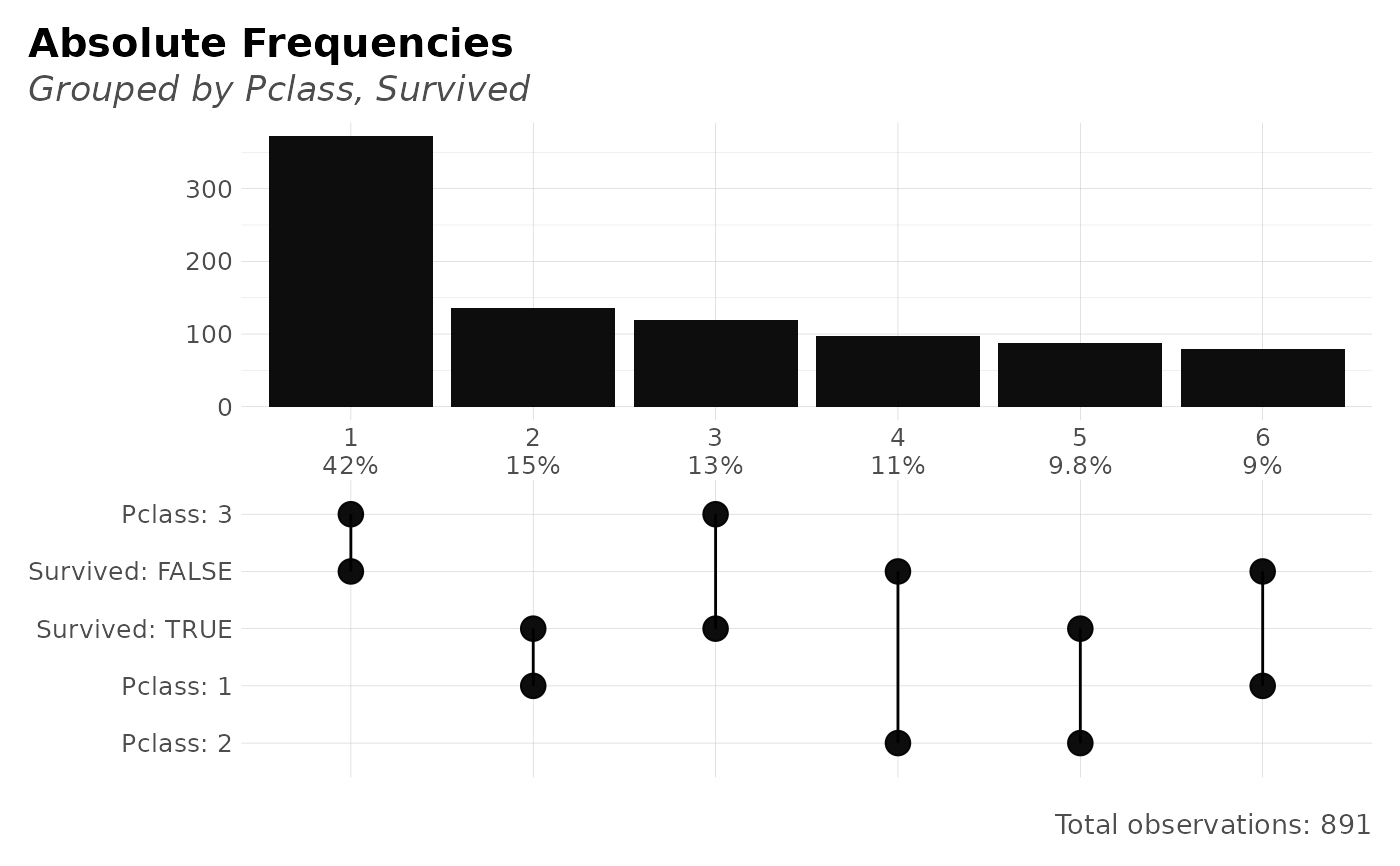
x <- freqs_plot(dft, Pclass, Survived)
x$data
#> # A tibble: 12 × 7
#> order n p pcum name value label
#> <chr> <int> <dbl> <dbl> <chr> <chr> <chr>
#> 1 6 80 8.98 100 Pclass 1 Pclass: 1
#> 2 6 80 8.98 100 Survived FALSE Survived: FALSE
#> 3 5 87 9.76 91.0 Pclass 2 Pclass: 2
#> 4 5 87 9.76 91.0 Survived TRUE Survived: TRUE
#> 5 4 97 10.9 81.3 Pclass 2 Pclass: 2
#> 6 4 97 10.9 81.3 Survived FALSE Survived: FALSE
#> 7 3 119 13.4 70.4 Pclass 3 Pclass: 3
#> 8 3 119 13.4 70.4 Survived TRUE Survived: TRUE
#> 9 2 136 15.3 57.0 Pclass 1 Pclass: 1
#> 10 2 136 15.3 57.0 Survived TRUE Survived: TRUE
#> 11 1 372 41.8 41.8 Pclass 3 Pclass: 3
#> 12 1 372 41.8 41.8 Survived FALSE Survived: FALSE
plot(x)
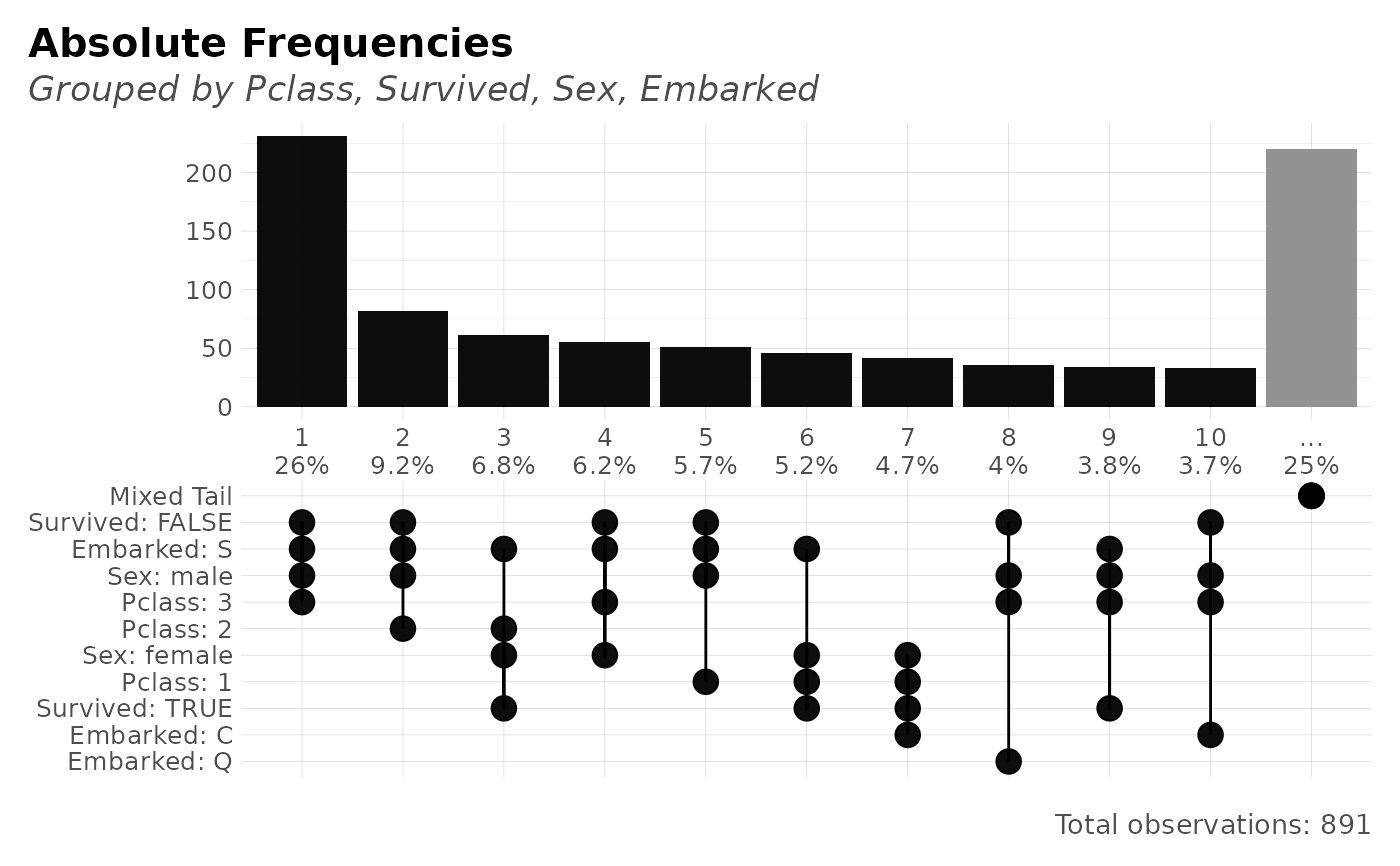
 freqs_plot(dft, Pclass, Survived, Sex, Embarked)
#> Showing 10 most frequent values. Tail of 22 other values grouped into one
freqs_plot(dft, Pclass, Survived, Sex, Embarked)
#> Showing 10 most frequent values. Tail of 22 other values grouped into one
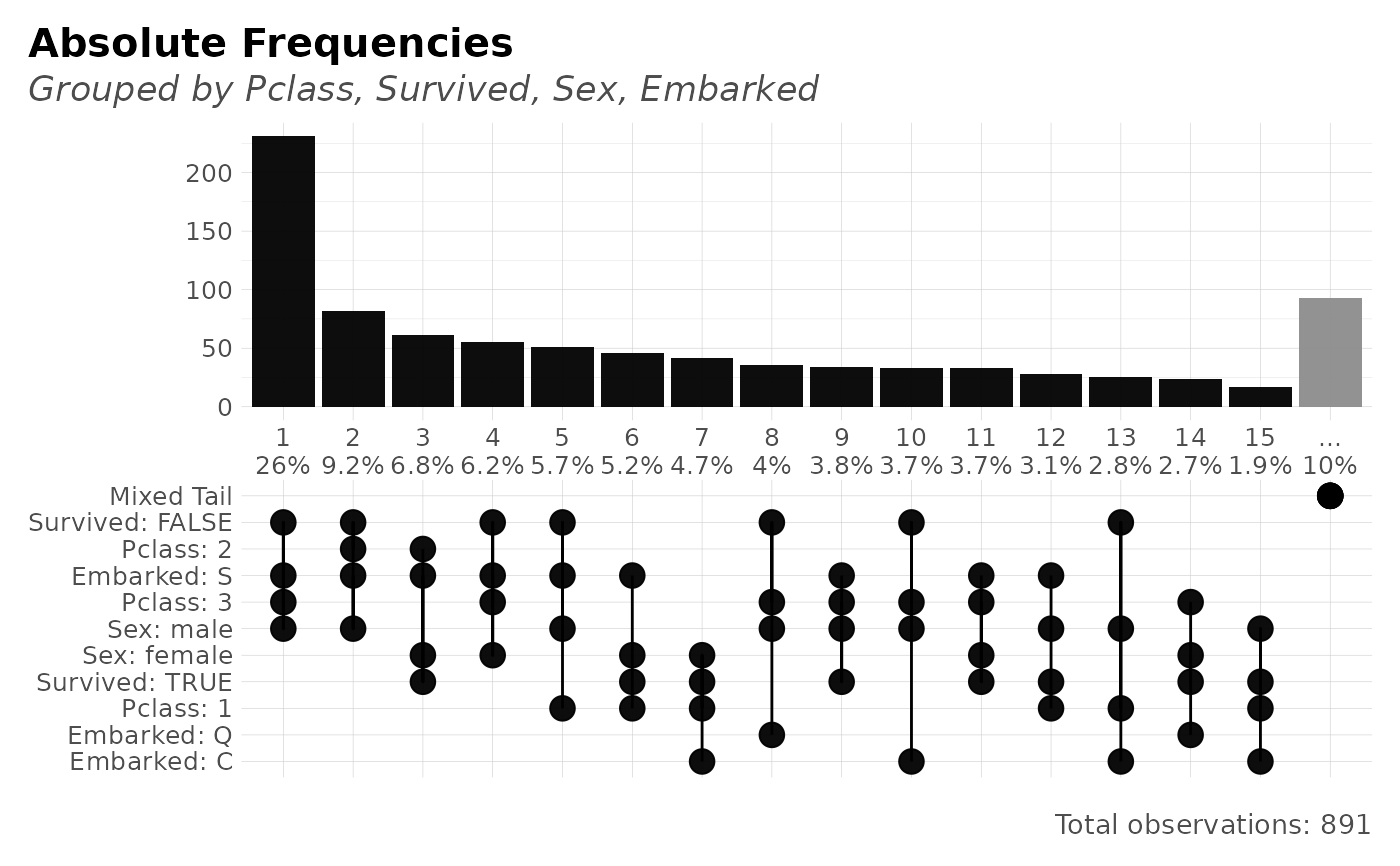
 freqs_plot(dft, Pclass, Survived, Sex, Embarked, top = 15)
#> Showing 15 most frequent values. Tail of 17 other values grouped into one
freqs_plot(dft, Pclass, Survived, Sex, Embarked, top = 15)
#> Showing 15 most frequent values. Tail of 17 other values grouped into one